| CCDC177 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | CCDC177, C14orf162, PLPL, coiled-coil domain containing 177 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 2686414 HomoloGene: 128326 GeneCards: CCDC177 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Coiled-Coil Domain Containing 177 (CCDC177) is a protein, which in humans, is encoded by the gene CCDC177.[5] It is composed of a coiled helical domain that spans half of the protein. CCDC177 deletions are associated with intellectual disability and congenital heart defects.[6]
Gene
The CCDC177 Gene is located on chromosome 14 at 14q24.1, and contains 2 exons.

The CCDC177 gene is part of the CCDC gene family, which encodes proteins involved in signal transduction and signal transcription.[7]
Other known aliases for the CCDC177 gene are Chromosome 14 Open Reading Frame 162 (C14orf162), and Myelin Proteolipid Protein-Like Protein (PLPL).[5]
mRNA transcripts
CCDC177 has 1 variant, which encodes Isoform 1 in humans. The mRNA sequence for this variant is 4,182 base pairs in length.[5] Both exons are present in the variant, however the coding region is entirely within Exon 2.
Protein
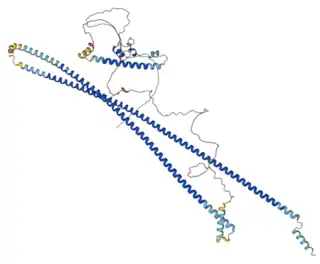
CCDC177 Isoform 1 in humans is 707 amino acids long[5] with a predicted molecular weight of 80 kDa.[9] It is rich in arginine, and glutamate, and poor in isoleucine relative to other proteins. The isoelectric point is 11.[10] The human protein is also rich in arginine-glutamate motifs, which are implicated in cell survival signaling.[11]
Domains and motifs
Humans CCDC177 includes one domain of unknown function (DUF4659), multiple disordered regions, and an alanine-rich motif.[5]
Structure
Proteins of the coiled coil domain containing (CCDC) family contain large coiled helical domains.[7][12] The coiled helical domain within the human CCDC177 protein fully overlaps the domain of unknown function (DUF4659).

Gene-level regulation
CCDC177 mRNA is ubiquitously expressed across adult human tissues, but is low in expression in fetal tissues throughout the body. It is also less abundant in immune cells such as B cells, T cells, and NK cells.[13]
Protein-level regulation
Sub-cellular location
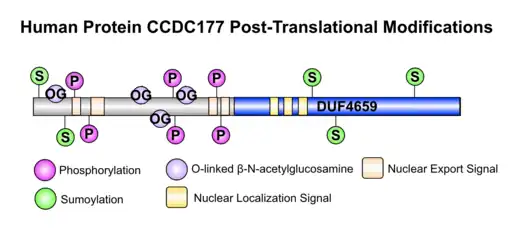
Human CCDC177 contains multiple nuclear localization signals, indicating that is found in the nucleus.[14] The protein also contains multiple nuclear export signals, indicating protein movement between the nucleus and cytosol.[15] The locations of the various kinases phosphorylating the CCDC177 protein implicate phosphorylation in CCDC177's movement between the nucleus and cytosol.[16]
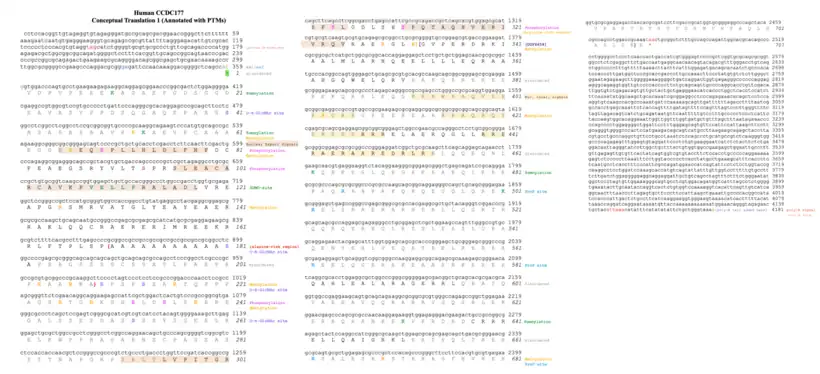
Post translational modifications
In CCDC177, phosphorylation and O-GlcNAc modifications are predicted to occur on several serine residues,[17] while SUMOylation occurs on select lysine residues.[18]
The types of kinases that phosphorylate highly conserved serine residues (conserved across current CCDC177 orthologs) in the CCDC177 protein sequence are located in the nucleus and cytosol. These kinases include Protein Kinase A which is located in the cytosol and nucleus,[19] Cyclin-dependent kinase 5 located in the cytosol,[20] and Protein Kinase C located in the nucleus.[21]
Conservation
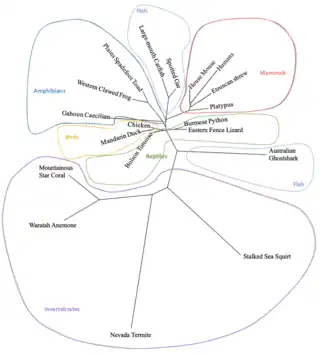
CCDC177 has no paralogs in humans. Orthologs are currently found in mammals, birds, reptiles, amphibians, fish, and invertebrates.[23]
| Organism ("Genus Species") | Common Name | Taxonomic Order | Median Date of Divergence (MYA) | Accession # | Sequence Length (aa) | Sequence Identity to Human Protein (%) | Sequence Similarity to Human Protein (%) | |
|---|---|---|---|---|---|---|---|---|
| Mammals | Homo sapiens | Human | Hominidae | 0 | NP_001258436.1 | 707 | 100 | 100 |
| Mus Musculus | Mouse | Rodentia | 87 | NP_001008423.2 | 706 | 90.6 | 94.1 | |
| Equus caballus | Horse | Perissodactyla | 94 | XP_023483759.1 | 700 | 93.9 | 95.3 | |
| Suncus etruscus | Etruscan Shrew | Eulipotyphla | 94 | XP_049626320.1 | 712 | 78.9 | 85.2 | |
| Phascolarctos cinereus | Koala | Marsupialia | 160 | XP_020860505.1 | 709 | 73 | 82 | |
| Ornithorhynchus anatinus | Platypus | Monotremata | 180 | XP_028920460.1 | 700 | 67.6 | 75.9 | |
| Birds | Gallu gallus | Chicken | Galliformes | 319 | XP_040527977.1 | 692 | 54.8 | 67.5 |
| Aix galericulata | Mandarin Duck | Anseriformes | 319 | KAI6068518.1 | 709 | 49.7 | 62.8 | |
| Reptiles | Sceloporus undulatus | Eastern Fence Lizard | Iguania | 319 | XP_042299999.1 | 710 | 52.9 | 68.8 |
| Gopherus flavomarginatus | Bolson Tortoise | Testudines | 319 | XP_050809463.1 | 714 | 51.7 | 65.3 | |
| Python bivittatus | Burmese Python | Serpentes | 319 | XP_007441661.1 | 740 | 49.0 | 62.8 | |
| Amphibians | Geotrypetes seraphini | Gaboon Caecilian | Gymnophiona | 352 | XP_033808243.1 | 663 | 47.6 | 63.2 |
| Spea bombifrons | Plains Spadefoot Toad | Anura | 352 | XP_053330589.1 | 688 | 41.1 | 58.9 | |
| Xenopus tropicalis | Western Clawed Frog | Anura | 352 | XP_002935376.2 | 679 | 40.6 | 58.6 | |
| Fish | Lepisosteus oculatus | Spotted Gar | Lepisosteiformes | 429 | XP_015206663.1 | 712 | 46.5 | 61.9 |
| Silurus meridionalis | Large-mouth Catfish | Siluriformes | 429 | KAI5102643.1 | 707 | 45.2 | 60.8 | |
| Callorhinchus milii | Australian Ghostshark | Chimaeriformes | 462 | XP_042189074.1 | 710 | 27.8 | 43.6 | |
| Invertebrates | Styela clava | Stalked Sea Squirt | Stolidobranchia | 596 | XP_039248961.1 | 678 | 21.7 | 38.9 |
| Actinia tenebrosa | Waratah Anemone | Actiniaria | 715 | XP_031562596.1 | 712 | 28.0 | 44.4 | |
| Orbicella faveolata | Mountainous Star Coral | Scleractinia | 715 | XP_020615800.1 | 718 | 25.2 | 40.9 |
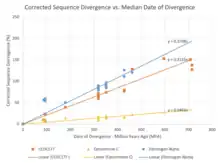
Rate of evolution
The protein encoded by CCDC177 evolves twice as fast as Cytochrome c and slightly slower than fibrinogen alpha, indicating that the CCDC177 gene has a moderately fast rate of evolution.
Interacting proteins
Human CCDC177 protein has notable interactions with the following proteins which are all associated with development and stem cell differentiation. All of the following proteins are located in the nucleus. These interactions implicate human CCDC177 in developmental processes and cell survival, and support its location in the nucleus.
- MYC binding protein 2 (MYCBP2) regulates neuronal growth and is required for proper axon growth.[26]
- Forkhead box protein N4 (FOXN4) is a transcription factor required for neural development and growth. It is especially important for specifying the fates of multipotent retinal progenitors.[27]
- Histone Deacetylase 5 (HDAC5) deacetylates lysine residues on the N-terminus tail of core histones and promotes cell cycle progression.[28]
- T-cell acute lymphocytic leukemia protein 1 (TAL1) is an oncogenic transcription factor in T-cell acute lymphoblastic leukemia, and is implicated in hematopoietic stem cell differentiation.[29]
Clinical significance
The CCDC177 gene can be utilized to develop prognostic tumor markers for neuroblastomas,[30] thyroid cancer,[31] and lung cancer.[32] CCDC177 is a methylation-driven gene in thyroid cancer, which was determined by examining proliferation and invasion of thyroid cancer (TC) cells in CCDC177 knockdown vectors. TC cells containing knockdown CCDC177 were highly proliferative and invasive.
Prognostic tumor methylation markers were discovered in human neuroblastoma as well.[33] 78 significantly differentially methylated regions were identified from 396 sequenced tumor profiles. Methylation-specific PCR assays were also developed to determine which regions accurately predict survival outcomes. 5 of the 78 assays, including one located in CCDC177, predicted event-free survival. CCDC177 mRNA is also integral to the accurate prediction of overall survival in lung squamous cell carcinoma(LUSC) patients.
Interstitial deletions of chromosome 14 at the location 14q24.1q24.3, which includes CCDC177, are linked to mild intellectual disability, congenital heart defects, and brachydactyly.[6]Haploinsufficiency in one or several of the deleted genes is the cause for the deletions.
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000267909 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000062961 - Ensembl, May 2017
- ↑ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- 1 2 3 4 5 6 "CCDC177 coiled-coil domain containing 177 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2023-12-16.
- 1 2 Oehl‐Jaschkowitz, Barbara; Vanakker, Olivier M.; De Paepe, Anne; Menten, Björn; Martin, Thomas; Weber, Georg; Christmann, Alexander; Krier, Romain; Scheid, Simone; McNerlan, Susan E.; McKee, Shane; Tzschach, Andreas (2014). "Deletions in 14q24.1q24.3 are associated with congenital heart defects, brachydactyly, and mild intellectual disability". American Journal of Medical Genetics Part A. 164 (3): 620–626. doi:10.1002/ajmg.a.36321. ISSN 1552-4825. PMID 24357125. S2CID 36417832.
- 1 2 Liu, Zhen; Yan, Weiwei; Liu, Shaohua; Liu, Zhan; Xu, Ping; Fang, Weiyi (2023-07-01). "Regulatory network and targeted interventions for CCDC family in tumor pathogenesis". Cancer Letters. 565: 216225. doi:10.1016/j.canlet.2023.216225. ISSN 0304-3835. PMID 37182638. S2CID 258683797.
- ↑ "AlphaFold Protein Structure Database". alphafold.ebi.ac.uk. Retrieved 2023-12-16.
- ↑ "SAPS < Sequence Statistics < EMBL-EBI". www.ebi.ac.uk. Retrieved 2023-12-16.
- ↑ Kozlowski, Lukasz P. "IPC - ISOELECTRIC POINT CALCULATION OF PROTEINS AND PEPTIDES". isoelectric.org. Retrieved 2023-12-16.
- ↑ Chandana, Thimmegowda; P. Venkatesh, Yeldur (2016-05-25). "Occurrence, Functions and Biological Significance of Arginine-Rich Proteins". Current Protein & Peptide Science. 17 (5): 507–516. doi:10.2174/1389203717666151201192348. PMID 26916156.
- ↑ Priyanka, Patra Priyadarshini; Yenugu, Suresh (2021). "Coiled-Coil Domain-Containing (CCDC) Proteins: Functional Roles in General and Male Reproductive Physiology". Reproductive Sciences. 28 (10): 2725–2734. doi:10.1007/s43032-021-00595-2. ISSN 1933-7191. PMID 33942254. S2CID 233487566.
- 1 2 "GDS596 / 220887_at". www.ncbi.nlm.nih.gov. Retrieved 2023-12-16.
- ↑ "Motif Scan". myhits.sib.swiss. Retrieved 2023-12-16.
- ↑ "LocNES NES prediction tool by Chook Lab". prodata.swmed.edu. Retrieved 2023-12-16.
- ↑ Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015 43:D512-20.
- ↑ "NetPhos 3.1 - DTU Health Tech - Bioinformatic Services". services.healthtech.dtu.dk. Retrieved 2023-12-16.
- ↑ "GPS-SUMO: Prediction of SUMOylation Sites & SUMO-interacting Motifs". sumo.biocuckoo.cn. Retrieved 2023-12-16.
- ↑ Klussmann, Enno (2007-01-01), Enna, S. J.; Bylund, David B. (eds.), "Protein Kinase A", xPharm: The Comprehensive Pharmacology Reference, New York: Elsevier, pp. 1–9, doi:10.1016/b978-008055232-3.60534-3, ISBN 978-0-08-055232-3, retrieved 2023-12-16
- ↑ Ino, H.; Chiba, T. (1996-09-02). "Intracellular localization of cyclin-dependent kinase 5 (CDK5) in mouse neuron: CDK5 is located in both nucleus and cytoplasm". Brain Research. 732 (1–2): 179–185. doi:10.1016/0006-8993(96)00523-9. ISSN 0006-8993. PMID 8891282. S2CID 20687258.
- ↑ Ringvold, H. C.; Khalil, R. A. (2017-01-01), Khalil, Raouf A. (ed.), "Chapter Six - Protein Kinase C as Regulator of Vascular Smooth Muscle Function and Potential Target in Vascular Disorders", Advances in Pharmacology, Vascular Pharmacology, Academic Press, vol. 78, pp. 203–301, doi:10.1016/bs.apha.2016.06.002, PMC 5319769, PMID 28212798, retrieved 2023-12-16
- ↑ "IBS - Database Visualization". ibs.biocuckoo.org. Retrieved 2023-12-16.
- ↑ "Protein BLAST: search protein databases using a protein query". blast.ncbi.nlm.nih.gov. Retrieved 2023-12-16.
- ↑ Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403-10. doi: 10.1016/S0022-2836(05)80360-2. PMID 2231712.
- ↑ "NGPhylogeny.fr". ngphylogeny.fr. Retrieved 2023-12-16.
- ↑ AlAbdi, Lama; Desbois, Muriel; Rusnac, Domniţa-Valeria; Sulaiman, Raashda A; Rosenfeld, Jill A; Lalani, Seema; Murdock, David R; Burrage, Lindsay C; Undiagnosed Diseases Network; Billie Au, Ping Yee; Towner, Shelley; Wilson, William G; Wong, Lawrence; Brunet, Theresa; Strobl-Wildemann, Gertrud (2023-04-19). "Loss-of-function variants in MYCBP2 cause neurobehavioural phenotypes and corpus callosum defects". Brain. 146 (4): 1373–1387. doi:10.1093/brain/awac364. ISSN 0006-8950. PMC 10319777. PMID 36200388.
- ↑ Islam, Mohammed M.; Li, Ying; Luo, Huijun; Xiang, Mengqing; Cai, Li (2013-11-15). "Meis1 regulates Foxn4 expression during retinal progenitor cell differentiation". Biology Open. 2 (11): 1125–1136. doi:10.1242/bio.20132279. ISSN 2046-6390. PMC 3828759. PMID 24244849.
- ↑ Wang, Yaomei; Li, Wei; Schulz, Vincent P.; Zhao, Huizhi; Qu, Xiaoli; Qi, Qian; Cheng, Yong; Guo, Xinhua; Zhang, Shijie; Wei, Xin; Liu, Donghao; Yazdanbakhsh, Karina; Hillyer, Christopher D.; Mohandas, Narla; Chen, Lixiang (2021-10-28). "Impairment of human terminal erythroid differentiation by histone deacetylase 5 deficiency". Blood. 138 (17): 1615–1627. doi:10.1182/blood.2020007401. ISSN 0006-4971. PMC 8554652. PMID 34036344.
- ↑ Hoang, T.; Lambert, J.A.; Martin, R. (2016), "SCL/TAL1 in Hematopoiesis and Cellular Reprogramming", Current Topics in Developmental Biology, Elsevier, 118: 163–204, doi:10.1016/bs.ctdb.2016.01.004, ISBN 978-0-12-803319-7, PMID 27137657, retrieved 2023-12-16
- ↑ Ram Kumar, Ram Mohan; Schor, Nina Felice (2018-04-24). "Methylation of DNA and chromatin as a mechanism of oncogenesis and therapeutic target in neuroblastoma". Oncotarget. 9 (31): 22184–22193. doi:10.18632/oncotarget.25084. ISSN 1949-2553. PMC 5955135. PMID 29774131.
- ↑ Chen, Zhiwei; Liu, Xiaoli; Liu, Fangfang; Zhang, Guolie; Tu, Haijian; Lin, Wei; Lin, Haifeng (2021-08-31). "Identification of 4-methylation driven genes based prognostic signature in thyroid cancer: an integrative analysis based on the methylmix algorithm". Aging. 13 (16): 20164–20178. doi:10.18632/aging.203338. ISSN 1945-4589. PMC 8436924. PMID 34456184.
- ↑ Ju, Qiang; Zhao, Yan-jie; Ma, Sai; Li, Xin-mei; Zhang, Heng; Zhang, Shao-qiang; Yang, Yuan-ming; Yan, Song-xia (2020). "Genome-wide analysis of prognostic-related lncRNAs, miRNAs and mRNAs forming a competing endogenous RNA network in lung squamous cell carcinoma". Journal of Cancer Research and Clinical Oncology. 146 (7): 1711–1723. doi:10.1007/s00432-020-03224-8. ISSN 0171-5216. PMID 32356177. S2CID 216650042.
- ↑ for the Children’s Cancer and Leukaemia Group (CCLG); Decock, Anneleen; Ongenaert, Maté; Cannoodt, Robrecht; Verniers, Kimberly; De Wilde, Bram; Laureys, Geneviève; Van Roy, Nadine; Berbegall, Ana P.; Bienertova-Vasku, Julie; Bown, Nick; Clément, Nathalie; Combaret, Valérie; Haber, Michelle; Hoyoux, Claire (2016). "Methyl-CpG-binding domain sequencing reveals a prognostic methylation signature in neuroblastoma". Oncotarget. 7 (2): 1960–1972. doi:10.18632/oncotarget.6477. ISSN 1949-2553. PMC 4811509. PMID 26646589.



